Quinoline Derivatives: In Vitro Antimicrobial Study
Sumitra Chanda, Pooja Moteriya , Paras Ramavat
Affiliation
- 1Department of Chemistry, Saurashtra University, Rajkot, India
- 2Department of Biosciences, Saurashtra University, Rajkot, India
Corresponding Author
Shipra Baluja, Department of Chemistry, Saurashtra University, Rajkot, 360005, Gujarat, India, E-mail: shipra_baluja@rediffmail.com
Citation
Baluja, S., et al. Quinoline Derivatives: In Vitro Antimicrobial Study. (2017) J Pharm Pharmaceutics 4(1): 105- 110.
Copy rights
© 2017 Baluja, S. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Quinoline derivatives; Dimethyl sulphoxide; N, N-dimethyl formamide; Gram positive bacteria; Gram negative bacteria; Fungal strains
Abstract
Some quinoline derivatives were synthesized and their structures were confirmed by IR, 1H NMR and mass spectroscopy. Screening of all these synthesized compounds were done in vitro against four Gram positive bacteria, four Gram negative bacteria and four fungal strains in dimethyl sulphoxide and N, N-dimethyl formamide. It is observed that N, N-dimethyl formamide is good solvent for the these compounds in selected strains and compounds containing cyano and nitro groups are more effective in inhibiting these strains.
Introduction
Heterocyclic compounds occupy a central position among those molecules that makes life possible. The majority of pharmaceuticals and biologically active agrochemicals are heterocyclic while countless additives and modifiers used in industrial applications ranging from cosmetics, reprography, information storage and plastics are heterocyclic in nature[1]. The presence of heterocycles in all kind of organic substances have much interest in biology, optics, pharmacology, biology, electronics, material sciences etc. Many natural drugs such as papaverine, theobromine, quinine, emetine, theophylline, atropine, procaine, codeine, reserpine and morphine[2-4] contain heterocyclic nucleus in their structure. Heterocycles are chemically more flexible and better able to respond to many demands of biochemical systems. The importance of heterocycles provides a significant basis for the development of new methods for their synthesis. Synthesis of various heterocyclic compounds with sulfur, nitrogen, oxygen, phosphorus and selenium have been reported[5-10].
Out of various classes of hetercycles, Quinolines have attracted much attention, because a large number of natural and synthetic products possess this heterocyclic unit[11-13]. Literature survey shows that various quinoline derivatives are known to possess a wide range of pharmacological properties, such as antiviral[14], antitubercular[15], antidiabetic[16], antibacterial[17], anticancer[18], antiarthritic, analgesic[19] antiinflammatory[20], antioxidant[21] etc.
Owing to their interesting biological properties, in the present work some novel quinoline derivatives were synthesized from arylidine, dimidone and ammonium acetate. The characterization of these synthesized compounds was done by IR, 1H NMR and mass spectral analysis.
The antimicrobial activity of the synthesized compounds was done against some pathogenic Gram positive and Gram negative bacteria and fungi in dimethyl sulphoxide and N, N-dimethyl formamide.
Experimental
Synthesis of arylidine
Equimolar methanolic solution of substituted aldehyde and malano nitrile using piperidine as a base was stirred for 30 minutes at room temperature. The progress of reaction was confirmed by Thin Layer Chromatography (TLC) (Performed on aluminium coated plate Gel60F254 (E.Merck) using (0.6: 0.4-Hexane: Ethyl acetate) as mobile phase. After the completion of reaction, the reaction mass was filtered, washed with methanol and dried under vacuum. The product was crystallized from methanol. Similarly, other substituted arylidines were synthesized.
Synthesis of quinoline derivatives
The methanolic solution of above synthesized substituted arylidine (1 mole), dimidone (1 mole) and ammonium acetate (5 mole) was stirred at room temperature for one hour. The progress of reaction was confirmed by Thin Layer Chromatography (TLC) (Performed on aluminium coated plate Gel60F254 (E.Merck)) using (0.5: 0.5-Hexane: Ethyl acetate) as mobile phase. The reaction mass was filtered, dried and was crystallized from methanol.
The scheme of synthesized compounds is given in (Figure 1).
Figure 1: Reaction scheme for the synthesis of compounds.
Structure confirmation
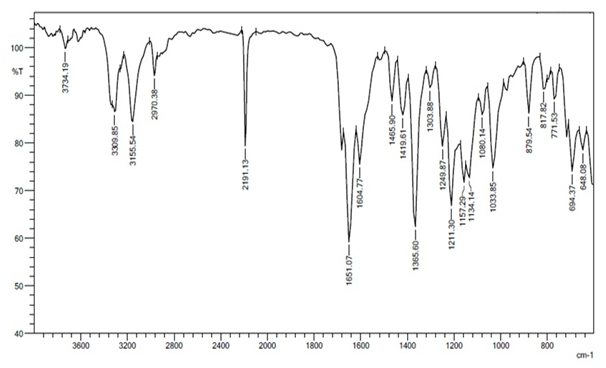
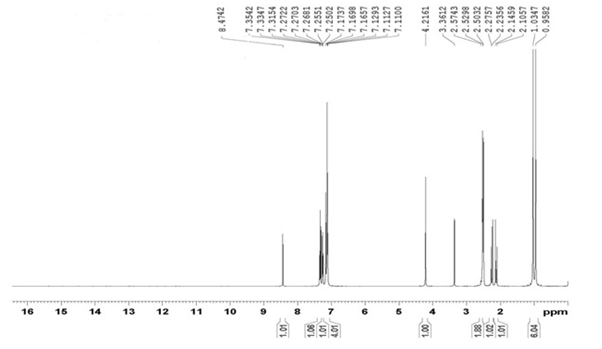
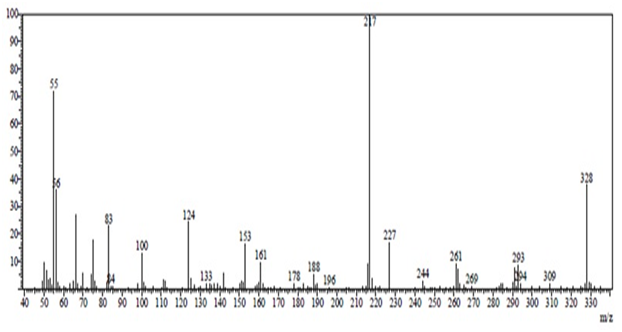
The structures of synthesized crystallized compounds were confirmed by FTIR, 1H NMR and mss spectral data. IR spectra were recorded on Shimadzu FT-IR-8400 instrument. 1H-NMR spectra were taken on a Bruker AVANCE II 400 using DMSO-d6 and mass spectra were determined using direct inlet probe on a GCMS-QP-2010 mass spectrometer. (Figures 2 - 4) shows IR, 1H NMR and mass spectra of R-1.
Figure 2: IR spectrum of compound R-1.
Figure 3: 1H NMR spectrum of compound R-1.
Figure 4: Mass spectrum of compound R-1.
Microorganisms tested
The studied microorganisms were obtained from National Chemical Laboratory (NCL), Pune, India. The microorganisms were maintained at 4°C. The Gram positive bacteria studied were Bacillus cereus ATCC11778 (BC); Staphylococcus aureus ATCC29737 (SA), Corynebacterium rubrum ATCC14898 (CR), Listeria monocytogenes ATCC19112 (LM), Gram negative bacteria were Escherichia coli NCIM2931 (EC), Pseudomonas aeruginosa ATCC27853 (PA), Salmonella typhimurium ATCC23564 (ST), Klebsiella pneumoniae NCIM2719 (KP) and fungal strains were Candida glabrata NCIM3448 (CG), Candida epicola NCIM3367 (CE), Candida albicans ATCC2091 (CA) and Cryptococcus neoformans NCIM3542 (CN). The microorganisms studied are clinically important ones causing several infections and food spoilage. In vitro antimicrobial activity of the quinoline was studied against pathogenic microbial strains by the agar well diffusion method[22].
Prediction of molecular descriptors and bioactivity score
Molinspiration software was used for calculation of molecular properties such as lipophilicity (Log P), total polar surface area, number of hydrogen bond doners and acceptors, number of atoms, number of rotatable bonds etc. Further, prediction of bioactivity score for drug targets such as G-protein coupled receptor (GPCR) ligands, kinase inhibitors, ion channel modulators, enzymes and nuclear receptors etc.
To determine drug likeness of compounds, Lipinski’s rule[23] is applied. It is a thumb rule of five to determine if a compound has properties that would make it likely to be drug. Lipinski’s rule states that absorption or permeation of a molecule is more likely when the molecular mass less than 500 daltons, the octanol-water partition coefficient (log P) is not greater than 5 and the molecule has not more than 5 hydrogen bond donors (nitrogen or oxygen atoms with one or more hydrogen atoms) and not more than 10 hydrogen bond acceptors (nitrogen or oxygen atoms).
Further, the probability of organic molecules to be drug like can also be predicted by their bioactivity score[24]. If the bioactivity score is greater than zero, compound is active, if score is in the range 0.0 to - 5.0 then compound is moderately active but less than - 5.0 score suggests inactive compound.
Results and Discussion
The physical constants of synthesized compounds are reported in (Table 1).
Table 1: The physical constants of all the synthesized compounds.
| Compound Code | Substitution R | M.F. | M.W. | Yield (%) | M.P.°C | Rf* value |
|---|---|---|---|---|---|---|
| R-1 | -3-Cl | C18H18ClN3O | 327 | 81 | 280 | 0.57 |
| R-2 | -2,5-di-OCH3 | C20H23N3O3 | 353 | 78 | 274 | 0.52 |
| R-3 | -4- CN | C19H18N4O | 318 | 82 | 292 | 0.58 |
| R-4 | -3-NO2 | C18H18N4O3 | 338 | 78 | 288 | 0.44 |
*0.5: 0.5-Hexane: Ethyl acetate
Spectral data
R-1
IR (cm-1): 1303.88 (C-H bend.), 1465.90 (C-C str.), 1495.90 (C=C str.), 1033.85 (C-H bend.), 1651.07 (-C=O str.), 3309.85 (-NH2 str.), 3155.59 (-NH str. ), 1365.60 (C-N str.), 2191.13 (-CN str.), 694.37 (C-Cl str.), (C-O str.), (N-O str.). 1H NMR (DMSO-d6) δ(ppm) : 0.958 (3H, singlet, -CH3), 1.034 (1H, singlet, -CH3), 2.105 - 2.272 (2H, doublet, -CH2), 2.503- 2.574 (2H, doublet, -CH2), 4.216 (1H, singlet, -CH), 7.110 - 7.112 (2H, doublet, Ar-CH), 7.169 (2H, singlet, -NH2), 7.250 - 7.255 (1H, doublet, Ar-CH), 7.315 - 7.354 (1H, triplet, Ar-CH), 8.640 (1H, singlet, Ar-CH). MS: (m/z) = 327.
R-2
IR (cm-1): 1303.86 (C-H bend.), 1496.74 (C-C str.), 1495.85 (C=C str.), 1033.75 (C-H bend.), 1651.02 (-C=O str.), 3309.55 (-NH2 str.), 3155.57 (-NH str.), 1365.30 (C-N str.), 2191.03 (-CN str.), 1026.13 (C-O str.). 1H NMR (DMSO-d6) δ(ppm) : : 0.974 (3H, singlet, -CH3), 1.041 (3H, singlet, -CH3), 2.050 - 2.275 (2H, doublet, -CH2), 2.428 - 2.573 (2H, doublet, -CH2), 3.649 (3H, singlet, -OCH3), 3.702 (3H, singlet, -OCH3), 4.441 (1H, singlet, -CH), 6.506 (2H, singlet, -NH2), 6.720-6.742 (2H, doublet, Ar-CH), 6.870 (1H, singlet, Ar-CH), 8.630 (1H, singlet, -NH). MS: (m/z) = 353
R-3
IR (cm-1): 1303.81 (C-H bend.), 1496.76 (C-C str.), 1495.59 (C=C str.), 1033.74 (C-H bend.), 1651.21 (-C=O str.), 3309.75 (-NH2 str.), 3155.29 (-NH str.), 1365.64 (C-N str.), 2191.27 (-CN str.). 1H NMR (DMSO-d6) δ(ppm) : 1.039 (3H, singlet, -CH3), 1.111 (1H, singlet, -CH3), 2.101-2.141 (1H, doublet, -CH), 2.240 - 2.280 (1H, doublet, -CH), 2.534 - 2.508 (2H, doublet, -CH2), 4.309 (1H, singlet, -CH), 7.163 (2H, singlet, -NH2), 7.361 - 7.381 (2H, doublet, Ar-CH), 7.765 - 7.785 (2H, doublet, Ar-CH), 8.351 (1H, singlet, -NH). MS: (m/z) = 318.
R-4
IR (cm-1): 1303.58 (C-H bend.), 1496.25 (C-C str.), 1495.87 (C=C str.), 1033.87 (C-H bend.), 1651.11 (-C=O str.), 3309.84 (-NH2 str.), 3155.39 (-NH str.), 1365.50 (C-N str.), 2191.10 (-CN str.), 1027.62 (N-O str.). 1H NMR (DMSO-d6) δ(ppm) : 0.974 (3H, singlet, -CH3), 1.051 (1H, singlet, -CH3), 2.107-2.147 (1H, doublet, -CH), 2.261-2.301 (1H, doublet, -CH), 4.438 (1H, singlet, -CH), 7.210 (2H, singlet, -NH2), 7.609 - 7.695 (2H, multiplet, Ar-CH), 8.000 (1H, singlet, Ar-CH), 8.410 (1H, singlet, Ar-CH), 8.410 (1H, singlet, -NH). MS: (m/z) = 338.
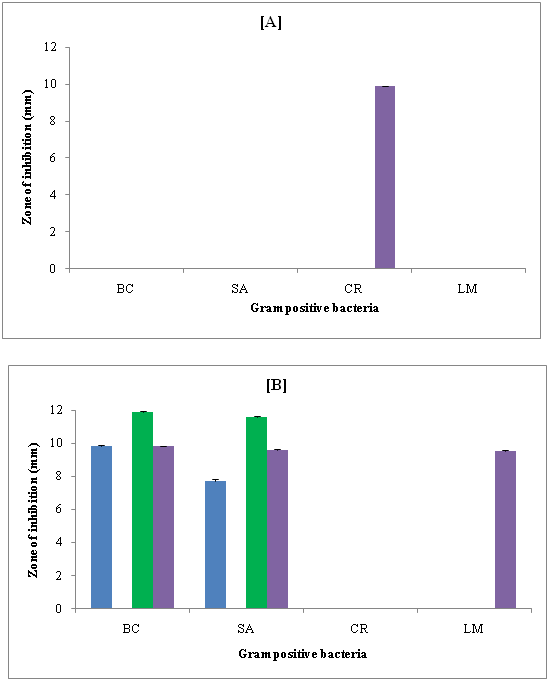
Figure 5: Zone of inhibition (mm) of compounds against Gram positive bacteria in [A] DMSO and [B] DMF
Antimicrobial activity: (Figure 5 A & B) shows the zone of inhibition of synthesized compounds against some Gram positive bacteria in dimethyl sulfoxide (DMSO) and N, N-dimethyl formamide (DMF) respectively. It is evident that none of the compound showed inhibition against Bacillus cereus (BC), Staphylococcus aureus (SA) and Listeria monocytogenes (LM) strains. Only R-4 exhibited inhibition against Corynebacterium rubrum (CR). Other compounds had no effect. However, in DMF, R-1, R-3 and R-4 showed inhibition against Bacillus cereus (BC) and Staphylococcus aureus (SA). None of the compound showed inhibition against Corynebacterium rubrum (CR). However, only R-3 could inhibit Listeria monocytogenes (LM).
The inhibition depends on solvent, structure and bacterial strain. All the synthesized compounds had the same central moiety but different substitution groups. (Table 1) shows the substitution group present in these compounds. Thus, 3-nitro group (present in R-4) was effective against Corynebacterium rubrum (CR) in DMSO. Other groups were not effective at all for all these Gram positive bacteria. However, in DMF, i.e., 3-chloro, 4-cyano and 3-nitro groups present in R-1, R-3 and R-4 respectively were effective against Bacillus cereus (BC) and Staphylococcus aureus (SA). For Listeria monocytogenes (LM), only R-3 containing 4-cyano group showed inhibition. However, none of these groups present in four compounds had any effect on Corynebacterium rubrum (CR).
Thus, R-2 had no effect against any of these bacterial strains in both DMF and DMSO. This compound contained 2, 4-dimethoxy group which was found to be not effective at all. Further, inhibition was higher in DMF as compared to DMSO against all the four selected Gram positive bacteria.
Thus, DMF was better solvent for the studied compounds. In DMSO, all the Gram positive bacteria except Corynebacterium rubrum (CR) were resistant whereas in DMF, Corynebacterium rubrum (CR) was most resistant bacteria.
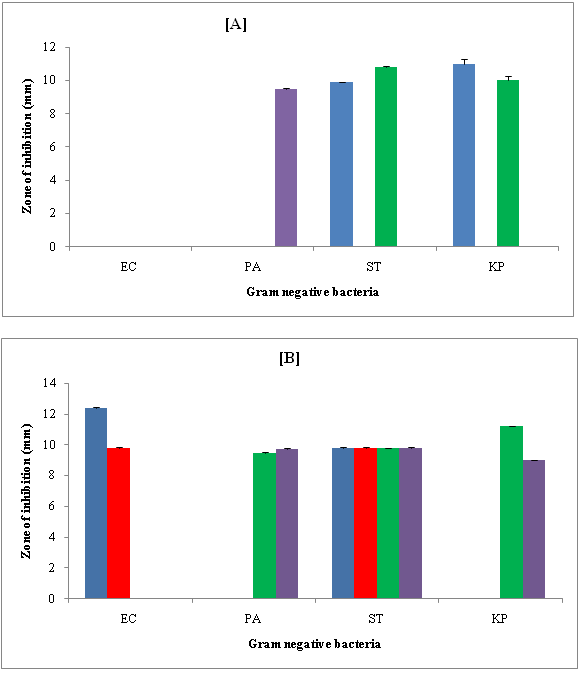
Figure 6: Zone of inhibition (mm) of compounds against Gram negative bacteria in [A] DMSO and [B] DMF.
(Figure 6 A & B) shows zone of inhibition of synthesized compounds against Gram negative bacteria in DMSO and DMF. In DMSO, against Escherichia coli (EC), none of the compound showed inhibition. Against Pseudomonas aeruginosa (PA), only R-4 exhibited inhibition. R-1 and R-3 are effective only against Salmonella typhimurium (ST) and Klebsiella pneumoniae (KP). Thus, 3-chloro (as in R-1) and 4-cyano (as in R-3) groups inhibit both Salmonella typhimurium (ST) and Klebsiella pneumonia (KP). Only 3-nitro group was effective against PA. Comparison of inhibition of R-1 and R-3 shows that in DMSO, R-3 exhibited maximum inhibition Salmonella typhimurium (ST) whereas reverse is true against Klebsiella pneumoniae (KP). R-2 had no effect at all in DMSO.
In DMF, only R-1 and R-2 showed inhibition against Escherichia coli (EC) and inhibition is higher for R-1. However, against Pseudomonas aeruginosa (PA) and Klebsiella pneumoniae (KP), only R-3 and R-4 exhibited inhibition. Inhibition was higher for R-4 against PA and R-3 against KP. All the four compounds were effective against Salmonella typhimurium (ST) to the sane extent. Thus, 3-chloro and 2, 5-dimethoxy groups had affect against Escherichia coli (EC) and Salmonella typhimurium (ST) whereas 3-nitro and 4-cyano groups could inhibit all the Gram negative bacteria except Escherichia coli (EC).
Against all these Gram negative bacterial strains, more compounds exhibited inhibition in DMF as compared to DMSO. Thus, DMF was better solvent for the studied compounds against these Gram negative bacteria.
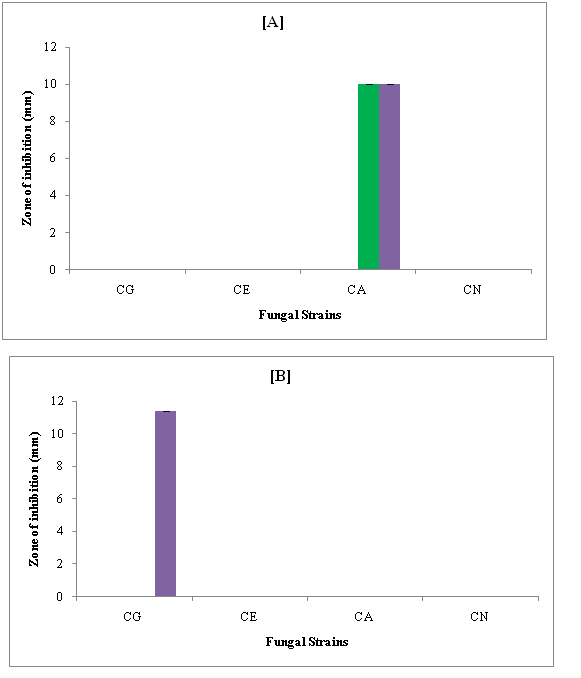
(Figure 7 A & B) show the zone of inhibition of compounds against some fungal strains in DMSO and DMF. It is evident that in DMSO, none of compounds showed inhibition against Candida glabrata (CG), Candida epicola (CE) and Cryptococcus neoformans (CN). Against Candida albicans (CA), inhibition is exhibited by only R-3 and R-4 to the same extent. Thus, 4-cyano and 3-nitro were effective against this strain to the same extent. In DMF, only R-4 containing 3-nitro group showed inhibition against Candida glabrata (CG). For other fungal strains, none of the compound was effective.
Figure 7: Zone of inhibition (mm) of compounds against fungal strains in [A] DMSO and [B] DMF.
Thus, Candida epicola (CE), Candida albicans (CA) and Cryptococcus neoformans (CN) were most resistant fungal strains and 3-nitro group was effective in both the solvents.
Drug likeness calculation on the basis of Lipinski rule: The molecular properties and bioactivity score of all synthesized compound calculated by molinspiration software are given in (Tables 2 & 3) respectively.
Table 2: Drug likeness score for compounds.
| Comp. Code | miLogP | TPSA | nAtoms | nON | nOHNH | n violation | n rotb. | volume |
|---|---|---|---|---|---|---|---|---|
| R-1 | 2.70 | 78.91 | 23 | 4 | 3 | 0 | 1 | 290.32 |
| R-2 | 1.61 | 97.38 | 26 | 6 | 3 | 0 | 3 | 327.88 |
| R-3 | 1.80 | 102.70 | 24 | 5 | 3 | 0 | 1 | 293.64 |
| R-4 | 1.98 | 124.74 | 25 | 7 | 3 | 0 | 2 | 300.12 |
Table 3: Bioactivity score of the compounds.
| Comp. Code | GPCR ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
|---|---|---|---|---|---|---|
| R-1 | -0.87 | -0.91 | -1.63 | -1.03 | -0.95 | -0.76 |
| R-2 | -0.83 | -0.91 | -1.51 | -0.85 | -0.89 | -0.70 |
| R-3 | -0.81 | -0.86 | -1.43 | -0.85 | -0.84 | -0.64 |
| R-4 | -0.97 | -0.91 | -1.61 | -1.02 | -0.97 | -0.80 |
All the synthesized compounds obeyed Lipinski’s rule. These compounds showed good permeability across cell membrane as miLog values were found below 5. TPSA of these compounds were found in the range of 78.91 - 124.74. Molecular weights of all the compounds were found to be less than 500 and number of hydrogen bond donors and hydrogen bond acceptors are 7 and 3 respectively for all the compounds. n violations is zero suggesting thereby that these compounds can be easily binded to receptor.
(Table 3) shows that all the parameters i.e., GPCR ligand, ion channel modulator, kinase inhibitor, nuclear receptor ligand, protease inhibitor, enzyme inhibitor were in the range (-1.63 to -0.64). This suggests that the synthesized compounds are moderately bioactive.
Conclusion
It is concluded that DMF was better solvent for antimicrobial activities of the studied compounds. Further, out of the four compounds, R-3 and R-4 containing 4-cyano and 3-nitro groups could inhibit more strains than 3-chloro and 2,5-di methoxy groups. The prediction of drug likeness of studied compounds also suggests moderate activity of compounds.
References
- 1. Dua, R., Shrivastava, S.., Sonwane, S.K., et al. Pharmacological Significance of Synthetic Heterocycles Scaffold: A Review. (2011) Adv Biol Res 5(3):120-144.
- 2. Chin, Y.W., Balunas, M.J., Chai, H.B., et al. Drug discovery from natural sources. (2006) AAPS J 8(2): E239-253.
- 3. Koehn, F.E., Carter, G.T. The evolving role of natural products in drug discovery. (2005) Nat Rev Drug Discov 4(3): 206-220.
- 4. Cordell, G.A., Quinn-Beattie, M.L., Farnsworth, N.R. The potential of alkaloids in drug discovery. (2001) Phytother Res 15(3): 183-205.
- 5. Kaur, N., Kishore, D. Erratum to Solid-phase synthesis of sulfur containing heterocycles. (2013) J Sulfur Chem 1-28.
- 6. Rakitin, O.A. One-pot synthesis of sulfur heterocycles from simple organic substrates. (2009) Arkivoc 129-149.
- 7. Trost, B.M., Lumb, J.P., Azzarelli, J.M. An atom-economic synthesis of nitrogen heterocycles from alkynes. (2011) J Am Chem Soc 133(4): 740-743.
- 8. Garibotto, F.M., Sortino, M.A., Kouznetsov, V.V., et al. Synthesis and antifungal activity of N-aryl-N-benzylamines and of their homoallyl analogues. (2011) Arkivoc 7: 149-161.
- 9. Reddy, P.V.G., Reddy, Y.B.K.., Reddy, C.S., et al. Synthesis and antimicrobial activity of novel phosphorus heterocycles with exocyclic PC link. (2004) Chem Pharm Bull 52(3): 307-310.
- 10. Abdel-Hafez, Sh.H. Selenium containing heterocycles: Synthesis, anti-inflammatory, analgesic and anti-microbial activities of some new 4-cyanopyridazine-3(2H)selenone derivatives. (2008) Eur J Med Chem 43(9): 1971-1977.
- 11. Nakayama, H., Kasoaka, Y. Chemical Identification of binding sites for calcium channel antagonists. (1996) Heterocycles 42: 901-909.
- 12. Lichitsky, B.V., Ivanov, S.N., Dudinov, S., et al. Reactions of cyclic enaminoketones with benzylidenemalononitriles. Synthesis of new fused heterocyclic systems containing the 1,4-dihydropyridine fragment. (2000) Russ Chem Bull International 50(12): 2428-2432.
- 13. Abdel-Gawad, S.M., El-Gaby, M.S.A., Heiba, H.I., et al. Synthesis and Radiation Stability of Some New Biologically Active Hydroquinoline and Pyrimido[4,5-b] quinoline Derivatives. (2005) J Chin Chem Soc 52(6): 1227-1236.
- 14. Zemtsova, M.N., Zimichev, A.V., Trakhtenberg, P.L., et al. Synthesis and antiviral activity of several quinoline derivatives. (2011) Pharma Chem J 45: 267.
- 15. Marcus, V.N.S., Karla, C.P., Carlos, R.K., et al. Synthesis and in vitro antitubercular activity of a series of quinoline derivatives. (2009) Bioorg Med Chem 17(4):1474-1480.
- 16. Srikanth, L., Raghunandan, N., Srinivas, P., et al. Synthesis and Evaluation of Newer Quinoline Derivatives of Thiazolidinediones for Their Antidiabetic Activity. (2010) Intern J Pharma BioSci. 1(4): 120-131.
- 17. Holla, B.S., Poojary, K..N., Poojary, B., et al. Synthesis, characterization and antibacterial activity studies on some fluorine containing quinoline-4-carboxylic acids and their derivatives. (2005) Indian J Chem 44B: 2114-2119.
- 18. Marganakop, S.B., Kamble, R.R., Hoskeri, J., et al. Facile synthesis of novel quinoline derivatives as anticancer agents. (2014) Med Chem Res 23(6): 2727-2735.
- 19. Clemence, F., Martret, O.L., Delevallee, F., et al. 4-Hydroxy-3-quinolinecarboxamides with antiarthritic and analgesic activities. (1988) J Med Chem 31(7): 1453-1462.
- 20. Calhoun, W., Carlson, R.P., Crossley, R. et al. Synthesis and Antiinflammatory Activity of Certain 5,6,7,8-Tetrahydroquinolines and Related Compounds. (1995) J Med Chem. 38(9): 1473-1481.
- 21. Prohaszka, L., Rozsnyal, T., Potentiation of the anticoccidial effect of salinomycin with dihydroquinoline-type antioxidants. (1990) Avian Pathol 19(1): 15-21.
- 22. Perez, C., Paul, M., Bazerque, P. An antibiotic assay by the agar well diffusion method. (1990) Acta Biol Med Exp: 113-115.
- 23. Lipinski, C.A., Lombardo, F., Dominy, B.W., et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. (2001) Adv Drug Deliv Rev 46(1-3): 3-26.
- 24. Verma, A. Lead finding from Phyllanthus debelis with hepatoprotective potentials. (2012) Asia- Pac J Trop Biomed 2(3): S1735-S1737.